Sorry for the late reply.
You’re right that rxd.Extracellular may be more expensive but you could easily include an outside region paralleling the cell (nrn_region=’o’) with minimal overhead cost. Additionally you can use a Parameter, so its value won’t be updated.
With a recent improvement on the development version (
https://github.com/neuronsimulator/nrn), which allows MultiCompartmentReaction sources (and destinations) to be on both the ‘i’ or ‘o’ side of the membrane, the pump mechanisms you described could be implemented as follows;
Code: Select all
import matplotlib.pyplot as plt
from neuron import h, rxd
from neuron.units import mV, ms, nM, mM
h.load_file('stdrun.hoc')
plt.ion()
soma = h.Section(name='soma')
soma.diam = soma.L = 10
PMCA_CONCENTRATION = 50 * nM
kf = 1e9 # forward rate [ca]*[pmca]*kf molecules/um**2/ms
kr, kcat = 1e5, 1e5 # reverse and catalytic rates [bound]*kr molecules/um**2/ms
cyt = rxd.Region(soma, name='cyt', nrn_region='i')
ecs = rxd.Region(soma, name='ecs', nrn_region='o', geometry=rxd.Shell(1, 2))
mem = rxd.Region(soma, name='mem', geometry=rxd.membrane())
ca = rxd.Species(cyt , d=1, name='ca', charge=2, initial=60*nM)
caecs = rxd.Parameter(ecs, name='ca', charge=2)
pcma = rxd.Species(mem, name='pcma', initial=PMCA_CONCENTRATION)
pcma_bound = rxd.Species(mem, name='pcma_bound', initial=0)
# define species on regions for the multicompartment reactions
cai, cao, unbound, bound = ca[cyt], caecs[ecs], pcma[mem], pcma_bound[mem]
# reversibly bind ca to the pump and generate a current
pcma_bind = rxd.MultiCompartmentReaction(cai + unbound, bound, kf, kr,
membrane_flux=True, membrane=mem)
# remove ca and returns the pump to the unbound state
# note: this reaction cannot generate a current because the extracellular
# (nrn_region='o') concentration (represented by a parameter) does not change.
pcma_extrue = rxd.MultiCompartmentReaction(bound, unbound + cao, kcat,
membrane=mem)
t = h.Vector().record(h._ref_t)
ca_vec = h.Vector().record(soma(0.5)._ref_cai)
unbound_vec = h.Vector().record(unbound.nodes._ref_value)
bound_vec = h.Vector().record(bound.nodes._ref_value)
v = h.Vector().record(soma(0.5)._ref_v)
h.finitialize(-65 * mV)
h.continuerun(100 * ms)
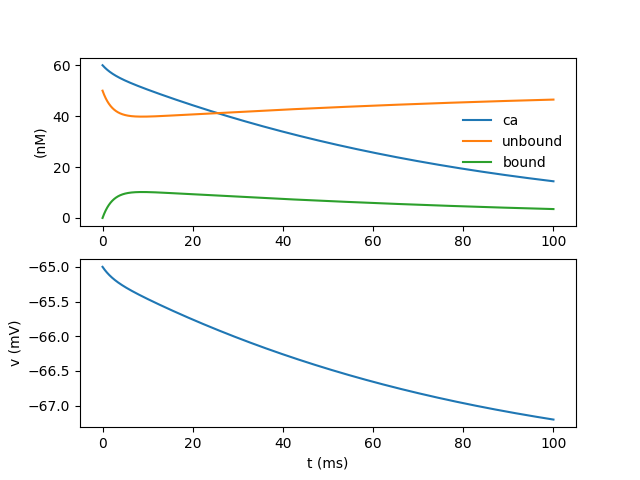
plt.subplot(2, 1, 1)
plt.plot(t, ca_vec / nM, label='ca')
plt.plot(t, unbound_vec / nM, label='unbound')
plt.plot(t, bound_vec / nM, label='bound')
plt.ylabel('(nM)')
plt.legend(frameon=False)
plt.subplot(2, 1, 2)
plt.plot(t, v)
plt.ylabel('v (mV)')
plt.xlabel('t (ms)')
plt.show()