2. In the NEURON Demonstrations window, click on the "Pyramidal" radio button.
3. Bring up the CellBuilder. This is a new CellBuilder, so it should only show a soma.
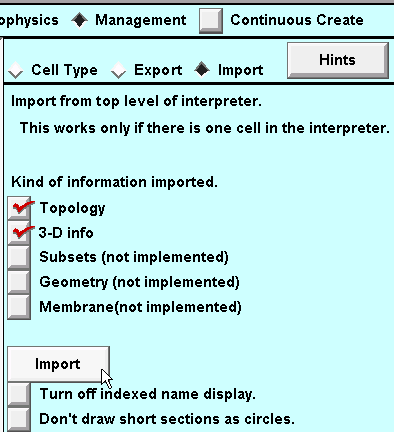
4. Select the Management page, then select the "Import" radio button.
5. Near the bottom of the CellBuilder is a button labeled "Import". Click on that button.
You will be warned that Import will discard whatever information is already in the CellBuilder, replacing it with a copy of information about the model cell that exists in the interpreter.
Click on "Go ahead and import".
Now the CellBuilder contains the pyramidal cell's topology and geometry.
Looks pretty messy.
Turning "indexed name display" off will clean things up a bit.
This is much clearer.
The CellBuilder now has all of the topology (branched architecture) and geometry (length and diameter) data of the detailed anatomical pyramidal cell model. We don't have to bother creating any sections ourselves, or specifying L or diam for any of them.
Save the CellBuilder to a session file (call it rawpyr.ses) and exit the demo program.
Then restart NEURON and retrieve rawpyr.ses. This reduces the chance of running into
"variable name conflicts".
Next we will create subsets that will be helpful when it comes time to specify biophysical properties.
Copyright © 1999-2006 by N.T. Carnevale and M.L. Hines, All Rights Reserved.